I've been looking at a lot of conformational manifolds lately. I used to do this with the RDKit's PyMol integration, but since that doesn't run on either of my laptops and since py3DMol looks awesome, I invested a bit of time in figuring out how to make it work with ipywidgets in jupyter. I figure this might be useful to others (and having the code online will be useful to me as well!), I'm doing another short post.
What I want to do is generate a set of conformers for a molecule and scroll through them interactively. Here's some code for doing that:
def drawit(m,p,confId=-1):
mb = Chem.MolToMolBlock(m,confId=confId)
p.removeAllModels()
p.addModel(mb,'sdf')
p.setStyle({'stick':{}})
p.setBackgroundColor('0xeeeeee')
p.zoomTo()
return p.show()
You use it like this:
import py3Dmol
from rdkit import Chem
from rdkit.Chem import AllChem
from ipywidgets import interact, interactive, fixed
m = Chem.MolFromSmiles(r'COc1ccc2[nH]c([S@@+]([O-])Cc3ncc(C)c(OC)c3C)nc2c1') # esomeprazole
m = Chem.AddHs(m)
AllChem.EmbedMultipleConfs(m,numConfs=10,randomSeed=0xf00d,useExpTorsionAnglePrefs=True,\
useBasicKnowledge=True)
# align to one of the ring systems:
AllChem.AlignMolConformers(m,m.GetSubstructMatch(Chem.MolFromSmarts('c1[nH]c2ccccc2n1')))
# now construct the view and interactive widget:
p = py3Dmol.view(width=400,height=400)
interact(drawit, m=fixed(m),p=fixed(p),confId=(0,m.GetNumConformers()-1));
And here's what the output looks like in the notebook:
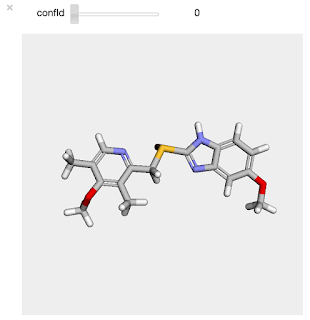
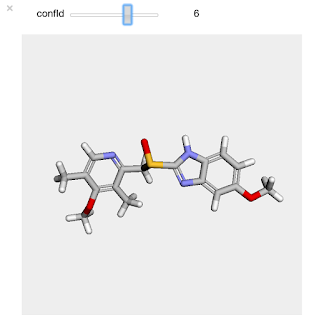
Simple and, I think, quite useful.
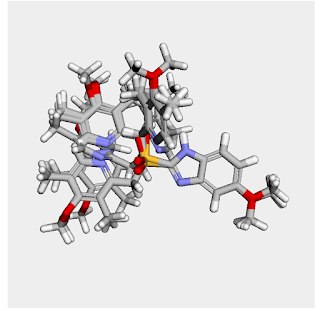
You can also display the whole bundle at once:
def drawit2(m,p,confId=-1):
mb = Chem.MolToMolBlock(m,confId=confId)
p.addModel(mb,'sdf')
p.setStyle({'stick':{}})
p.setBackgroundColor('0xeeeeee')
p.zoomTo()
p = py3Dmol.view(width=400,height=400)
for confId in range(10):
drawit2(m,p,confId)
p.show()
This yields:

1 comment:
Greg, unfortunately, the interactive slider bar example here doesn't seem to work in 2017.09 anymore. :-(
Some internal miscommunication due to the inclusion of py3Dmol being used internally in RDKit iPython now?
Post a Comment